7 Analysis
7.1 srvyr package
“The srvyr package aims to add dplyr like syntax to the survey package.” It is a very useful package for a variety of aggregations of survey data.
###makes some additions.
library(tidyverse)
library(srvyr)
library(kableExtra)
library(readxl)
df<-read_csv("inputs/UKR2007_MSNA20_HH_dataset_main_rcop.csv")
main_dataset <- read.csv("inputs/UKR2007_MSNA20_HH_dataset_main_rcop.csv", na.strings = "")
loop_dataset <- read.csv("inputs/UKR2007_MSNA20_HH_dataset_loop_rcop.csv", na.strings = "")
main_dataset <- main_dataset %>% select_if(~ !(all(is.na(.x)) | all(. == "")))
questions <- read_xlsx("inputs/UKR2007_MSNA20_HH_Questionnaire_24JUL2020.xlsx",sheet="survey")
choices <- read_xlsx("inputs/UKR2007_MSNA20_HH_Questionnaire_24JUL2020.xlsx",sheet="choices")
dfsvy<-as_survey(df)7.1.1 Categorical variables
srvyr package allows categorical variables to be broken down using a similar syntax as dplyr. Using dplyr you might typically calculate a percent mean as follows:
df %>%
group_by(b9_hohh_marital_status) %>%
summarise(
n=n()
) %>%
ungroup() %>%
mutate(
pct_mean=n/sum(n)
)## # A tibble: 6 × 3
## b9_hohh_marital_status n pct_mean
## <chr> <int> <dbl>
## 1 divorced 183 0.113
## 2 married 677 0.419
## 3 separated_married_but_not_living_together 37 0.0229
## 4 single 129 0.0798
## 5 unmarried_but_living_together 96 0.0594
## 6 widowed 495 0.306To calculate the percent mean of a categorical variable using srvyr object is required. The syntax is quite similar to dplyr, but a bit less verbose. By specifying the vartype as “ci” we also get the upper and lower confidence intervals
## # A tibble: 6 × 4
## b9_hohh_marital_status pct_mean pct_mean_low pct_mean_upp
## <chr> <dbl> <dbl> <dbl>
## 1 divorced 0.113 0.0977 0.129
## 2 married 0.419 0.395 0.443
## 3 separated_married_but_not_living_together 0.0229 0.0156 0.0302
## 4 single 0.0798 0.0666 0.0930
## 5 unmarried_but_living_together 0.0594 0.0478 0.0709
## 6 widowed 0.306 0.284 0.329To calculate the weigthed percent mean of a multiple response option you need to created a srvyr object including the weights. The syntax is similar to dyplr and allows for the total columns
weighted_object <- main_dataset %>% as_survey_design(ids = X_uuid, weights = stratum.weight)
weighted_table <- weighted_object %>%
group_by(adm1NameLat) %>% #group by categorical variable
summarise_at(vars(starts_with("b10_hohh_vulnerability.")), survey_mean) %>% #select multiple response question
ungroup() %>%
bind_rows(
summarise_at(weighted_object,
vars(starts_with("b10_hohh_vulnerability.")), survey_mean) # bind the total
) %>%
mutate_at(vars(adm1NameLat), ~replace(., is.na(.), "Total")) %>%
select(., -(ends_with("_se"))) %>% #remove the colums with the variance type calculations
mutate_if(is.numeric, ~ . * 100) %>%
mutate_if(is.numeric, round, 2)
print(weighted_table)## # A tibble: 3 × 11
## adm1NameLat b10_hohh_vulnerability.no b10_hohh_vulnerability… b10_hohh_vulner…
## <chr> <dbl> <dbl> <dbl>
## 1 Donetska 30.0 9.7 8.88
## 2 Luhanska 15.2 9.88 9.63
## 3 Total 26.4 9.74 9.07
## # … with 7 more variables: b10_hohh_vulnerability.veteran_of_war_ato <dbl>,
## # b10_hohh_vulnerability.single_parent <dbl>,
## # b10_hohh_vulnerability.family_with_3_or_more_children <dbl>,
## # b10_hohh_vulnerability.family_with_foster_children <dbl>,
## # b10_hohh_vulnerability.other_specify <dbl>,
## # b10_hohh_vulnerability.chronic_illness <dbl>,
## # b10_hohh_vulnerability.older_person <dbl>7.1.2 Numeric variables
srvyr treats the calculation/aggregation of numeric variables differently in an attempt to mirror dplyr syntax
to calculate the mean and median expenditure in dplyr you would likely do the following
df %>%
summarise(
mean_expenditure= mean(n1_HH_total_expenditure,na.rm=T),
median_expenditure=median(n1_HH_total_expenditure,na.rm=T),
)## # A tibble: 1 × 2
## mean_expenditure median_expenditure
## <dbl> <dbl>
## 1 5129. 4000If you wanted to subset this by another variable in dplyr you would add the group_by argument
df %>%
group_by(strata) %>%
summarise(
mean_expenditure= mean(n1_HH_total_expenditure,na.rm=T),
median_expenditure=median(n1_HH_total_expenditure,na.rm=T),
)## # A tibble: 4 × 3
## strata mean_expenditure median_expenditure
## <chr> <dbl> <dbl>
## 1 20km_rural 5488. 4000
## 2 20km_urban 6208. 5000
## 3 5km_rural 3919. 3075
## 4 5km_urban 4942. 4000This is the reason why the syntax also varies between categorical and numeric variables in srvyr. Therefore, to do the same using srvyr you would do the following (with a survey object). Note that due to this difference in syntax the na.rm argument works for numeric variables, but does not work for categorical variables. This was modified when srvyr was updated from v 0.3.8
## # A tibble: 1 × 3
## mean mean_low mean_upp
## <dbl> <dbl> <dbl>
## 1 5129. 4891. 5367.similar to dplyr you can easily add a group_by argument to add a subset calculation
dfsvy %>%
group_by(strata) %>%
summarise(
mean= survey_mean(n1_HH_total_expenditure,na.rm=T,vartype = "ci"),
)## # A tibble: 4 × 4
## strata mean mean_low mean_upp
## <chr> <dbl> <dbl> <dbl>
## 1 20km_rural 5488. 4980. 5996.
## 2 20km_urban 6208. 5661. 6756.
## 3 5km_rural 3919. 3469. 4370.
## 4 5km_urban 4942. 4604. 5280.7.2 Analysis with numerical variables
7.2.2 Median
####Spatstat Spatstat - library with set of different functions for analyzing Spatial Point Patterns but also quite useful for analysis of weighted data.
At first let’s select all numerical variables from the dataset using Kobo questionnaire and dataset. It can be done with the following custom function:
## Loading required package: spatstat.data## Loading required package: spatstat.geom## spatstat.geom 3.0-3##
## Attaching package: 'spatstat.geom'## The following object is masked from 'package:maditr':
##
## shift## The following object is masked from 'package:grid':
##
## as.mask## Loading required package: spatstat.random## spatstat.random 3.0-1## Loading required package: spatstat.explore## Loading required package: nlme##
## Attaching package: 'nlme'## The following object is masked from 'package:dplyr':
##
## collapse## spatstat.explore 3.0-5## Loading required package: spatstat.model## Loading required package: rpart## spatstat.model 3.0-2##
## Attaching package: 'spatstat.model'## The following object is masked from 'package:expss':
##
## valid## Loading required package: spatstat.linnet## spatstat.linnet 3.0-3##
## spatstat 3.0-2
## For an introduction to spatstat, type 'beginner'select_numerical <- function(dataset, kobo){
kobo_questions <- kobo[grepl("integer|decimal|calculate", kobo$type),c("type","name")]
names.use <- names(dataset)[(names(dataset) %in% as.character(kobo_questions$name))]
numerical <- dataset[,c(names.use,"X_uuid",'strata','stratum.weight')] #Here we can select any other relevant variables
numerical[names.use] <- lapply(numerical[names.use], gsub, pattern = 'NA', replacement = NA)
numerical[names.use] <- lapply(numerical[names.use], as.numeric)
return(numerical)
}
numerical_questions <- select_numerical(main_dataset, questions)
numerical_classes <- sapply(numerical_questions[,1:c(ncol(numerical_questions)-3)], class) #Here we can check class of each selected variable
numerical_classes <- numerical_classes["character" %in% numerical_classes] #and here we check if any variable has class "character"
numerical_questions <- numerical_questions[ , !names(numerical_questions) %in% numerical_classes] #if any variable has a character class then we remove it
rm(numerical_classes)#and here we removing vector with classes from our environmentNow let’s calculate weighted median for “n1_HH_total_expenditure”.
weighted.median(numerical_questions$n1_HH_total_expenditure, numerical_questions$stratum.weight, na.rm=TRUE)## [1] 4150But if we want to calculate weighted medians for each variable we will need to iterate this function on those variables. But first we will need to exclude variables with less than 3 observations.
counts <- numerical_questions %>%
select(-X_uuid, -strata) %>%
summarise(across(.cols = everything(), .fns= ~sum(!is.na(.)) ))%>%
t()#Calculating count of observation for each variable
numerical_questions <- numerical_questions[ , (names(numerical_questions) %in% rownames(subset(counts, counts[,1] > 3)))]
#removing variables with less than 3 observations
medians <- lapply(numerical_questions[1:46], weighted.median, w = numerical_questions$stratum.weight, na.rm=TRUE)%>%
as.data.frame()Now we can transpond resulting vector and add description to the calculation
medians <- as.data.frame(t(medians),stringsAsFactors = FALSE)
names(medians)[1] <- "Median_wght"
head(medians)## Median_wght
## b15_hohh_income 2780.0
## hohh_age_18 0.0
## age_18 0.0
## b21_num_additional_hh_members 0.5
## age_5_18_number 0.0
## age_5_12_number 0.07.3 Ratios
This function computes the % of answers for select multiple questions.
The output can be used as a data merge file for indesign.
The arguments are: dataset, disaggregation variable, question to analyze, and weight column.
7.3.1 Example1:
Question: e11_items_do_not_have_per_hh
Disaggregation variable: b9_hohh_marital_status
weights column: stratum.weight
ratios_select_multiple <-
function(df, x_var, y_prefix, weights_var) {
df %>%
group_by({
{
x_var
}
}) %>% filter(!is.na(y_prefix)) %>%
summarize(across(starts_with(paste0(y_prefix, ".")),
list(pct = ~ weighted.mean(., {
{
weights_var
}
}, na.rm = T))))
}
res <-
ratios_select_multiple(
main_dataset,
b9_hohh_marital_status,
"e11_items_do_not_have_per_hh",
stratum.weight
)
head(res)## # A tibble: 6 × 5
## b9_hohh_marital_status e11_items_do_no… e11_items_do_no… e11_items_do_no…
## <chr> <dbl> <dbl> <dbl>
## 1 divorced 0.0851 0.0859 0.393
## 2 married 0.0496 0.0545 0.409
## 3 separated_married_but_not_… 0.0138 0.0513 0.443
## 4 single 0.0726 0.0516 0.457
## 5 unmarried_but_living_toget… 0.0874 0.101 0.293
## 6 widowed 0.106 0.0923 0.504
## # … with 1 more variable:
## # e11_items_do_not_have_per_hh.hh_has_all_items_pct <dbl>7.3.2 Example2 - Analyzing multiple questions and combine the output:
Question: e11_items_do_not_have_per_hh and b11_hohh_chronic_illness
Disaggregation variable: b9_hohh_marital_status
weights column: stratum.weight
res <-
do.call("full_join", map(
c("e11_items_do_not_have_per_hh", "b11_hohh_chronic_illness"),
~ ratios_select_multiple(main_dataset, b9_hohh_marital_status, .x, stratum.weight)
))## Joining, by = "b9_hohh_marital_status"## # A tibble: 6 × 18
## b9_hohh_marital_status e11_items_do_no… e11_items_do_no… e11_items_do_no…
## <chr> <dbl> <dbl> <dbl>
## 1 divorced 0.0851 0.0859 0.393
## 2 married 0.0496 0.0545 0.409
## 3 separated_married_but_not_… 0.0138 0.0513 0.443
## 4 single 0.0726 0.0516 0.457
## 5 unmarried_but_living_toget… 0.0874 0.101 0.293
## 6 widowed 0.106 0.0923 0.504
## # … with 14 more variables:
## # e11_items_do_not_have_per_hh.hh_has_all_items_pct <dbl>,
## # b11_hohh_chronic_illness.blood_pressure_diseases_pct <dbl>,
## # b11_hohh_chronic_illness.cardiovascular_disease_pct <dbl>,
## # b11_hohh_chronic_illness.diabetes_need_insulin_pct <dbl>,
## # b11_hohh_chronic_illness.diabetes_does_not_need_insulin_pct <dbl>,
## # b11_hohh_chronic_illness.chronic_respiratory_condition_pct <dbl>, …7.3.3 Example3 - No dissagregation is needed and/or the data is not weighted:
Question: e11_items_do_not_have_per_hh and b11_hohh_chronic_illness
Disaggregation variable: NA
weights column: NA
main_dataset$all <- "all"
main_dataset$no_weights <- 1
res <-
do.call("full_join", map(
c("e11_items_do_not_have_per_hh", "b11_hohh_chronic_illness"),
~ ratios_select_multiple(main_dataset, all, .x, no_weights)
))## Joining, by = "all"## # A tibble: 1 × 18
## all e11_items_do_not_hav… e11_items_do_no… e11_items_do_no… e11_items_do_no…
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 all 0.0804 0.0779 0.417 0.515
## # … with 13 more variables:
## # b11_hohh_chronic_illness.blood_pressure_diseases_pct <dbl>,
## # b11_hohh_chronic_illness.cardiovascular_disease_pct <dbl>,
## # b11_hohh_chronic_illness.diabetes_need_insulin_pct <dbl>,
## # b11_hohh_chronic_illness.diabetes_does_not_need_insulin_pct <dbl>,
## # b11_hohh_chronic_illness.chronic_respiratory_condition_pct <dbl>,
## # b11_hohh_chronic_illness.musculoskeletal_system_and_joints_pct <dbl>, …7.4 Weights
Weights can be used for different reasons. In most of cases, we will use weights to correct for difference between strata size. Once the population has a certain (big) size, for the same design, the size of the sample won’t change much. In the case of the dataset, there are 4 stratas:
- within 20km of the border in rural areas : 89,408 households
- within 20km of the border in urban areas : 203,712 households
- within 20km of the border in rural areas : 39,003 households
- within 20km of the border in rural areas : 211,857 households
Using any sampling calculator, for a 95% confindence level, 5% margin of error and 5% buffer, we will have around 400 samples per strata even though the urban areas are much bigger.
Note: The error of 5% for 90,000 is 4,500 households while for 200,000 households is 10,000 households
If we look at each strata and the highest level of education completed by the household head (b20_hohh_level_education), we can see that the percentage of household were the head of household completed higher education varies between between 7 to 13 % in each strata.
## # A tibble: 4 × 5
## # Groups: strata [4]
## strata b20_hohh_level_education n tot_n prop
## <chr> <chr> <int> <int> <dbl>
## 1 20km_rural complete_higher 40 407 10
## 2 20km_urban complete_higher 51 404 13
## 3 5km_rural complete_higher 38 402 9
## 4 5km_urban complete_higher 28 404 7However, if we want to know overall the percentage of who finished higher education we cannot just take the overall percentages, i.e. \(\frac{40 + 51 +38 + 28}{407 + 404 + 402 + 404}\) = \(\frac{157}{1617}\) = 10%.
We cannot do this because the first strata represent 90,000 households, the second 200,000 households, the third 40,000 households and the last one 210,000 households. We will use weights to compensate this difference.
We will use this formula:
weight of strata = \(\frac{\frac{strata\\ population}{total\\ population}}{\frac{strata\\ sample}{total\\ sample}}\)
7.4.1 tidyverse
The following example will show how to calculate the weights with tidyverse.
To calculate weights, we will need all the following information: - population of the strata,
- total population,
- sample and
- total sample.
The population information should come from the sampling frame that was used to draw the sampling.
my_sampling_frame <- readxl::read_excel("inputs/UKR2007_MSNA20_GCA_Weights_26AUG2020.xlsx",
sheet = "for_r") %>%
rename(population_strata = population)
my_sampling_frame## # A tibble: 4 × 2
## strata population_strata
## <chr> <dbl>
## 1 20km_rural 89408
## 2 20km_urban 230712
## 3 5km_rural 39003
## 4 5km_urban 211857Then, we need to get the actual sample from the dataset.
sample_per_strata_table <- main_dataset %>%
group_by(strata) %>%
tally() %>%
rename(strata_sample = n) %>%
ungroup()
sample_per_strata_table## # A tibble: 4 × 2
## strata strata_sample
## <chr> <int>
## 1 20km_rural 407
## 2 20km_urban 404
## 3 5km_rural 402
## 4 5km_urban 404Then, we can join the tables together and calculate the weights per strata.
weight_table <- sample_per_strata_table %>%
left_join(my_sampling_frame) %>%
mutate(weights = (population_strata/sum(population_strata))/(strata_sample/sum(strata_sample)))
weight_table## # A tibble: 4 × 4
## strata strata_sample population_strata weights
## <chr> <int> <dbl> <dbl>
## 1 20km_rural 407 89408 0.622
## 2 20km_urban 404 230712 1.62
## 3 5km_rural 402 39003 0.275
## 4 5km_urban 404 211857 1.49Then we can finally add them to the dataset.
main_dataset <- main_dataset %>%
left_join(select(weight_table, strata, weights), by = "strata")
head(main_dataset$weights)## [1] 0.2747648 0.2747648 0.6221156 0.6221156 0.6221156 0.6221156We can check that each strata has only 1 weight.
## # A tibble: 4 × 3
## # Groups: strata [4]
## strata weights n
## <chr> <dbl> <int>
## 1 20km_rural 0.622 407
## 2 20km_urban 1.62 404
## 3 5km_rural 0.275 402
## 4 5km_urban 1.49 404We can check that the sum of weights is equal to the number of interviews.
## [1] TRUE7.4.2 surveyweights
surveyweights was created to calculate weights. The function weighting_fun_from_samplingframe creates a function that calculates weights from the dataset and the sampling frame.
Note: surveyweights can be found on github.
First you need to create the weighting function. weighting_fun_from_samplingframe will take 5 arguments:
- sampling.frame: a data frame with your population figures and stratas.
- data.stratum.column : column name of the strata in the dataset.
- sampling.frame.population.column : column name of population figures in the sampling frame.
- sampling.frame.stratum.column : column name of the strata in the sampling frame.
- data : dataset
##
## Attaching package: 'surveyweights'## The following object is masked from 'package:hypegrammaR':
##
## combine_weighting_functionsmy_weigthing_function <- surveyweights::weighting_fun_from_samplingframe(sampling.frame = my_sampling_frame,
data.stratum.column = "strata",
sampling.frame.population.column = "population_strata",
sampling.frame.stratum.column = "strata",
data = main_dataset)See that the my_weigthing_function is not a vector of weights. It is a function that takes the dataset as argument and returns a vector of weights.
## [1] TRUE## function (df)
## {
## if (!is.data.frame(df)) {
## stop("df must be a data.frame")
## }
## df <- as.data.frame(df, stringsAsFactors = FALSE)
## df <- lapply(df, function(x) {
## if (is.factor(x)) {
## return(as.character(x))
## }
## x
## }) %>% as.data.frame(stringsAsFactors = FALSE)
## if (!all(data.stratum.column %in% names(df))) {
## stop(paste0("data frame column '", data.stratum.column,
## "'not found."))
## }
## df[[data.stratum.column]] <- as.character(df[[data.stratum.column]])
## sample.counts <- stratify.count.sample(data.strata = asdhk[[data.stratum.column]],
## sf.strata = population.counts)
## if ("weights" %in% names(df)) {
## warning("'weights' is used as a column name (will not be calculated from the sampling frame)")
## }
## if (!all(names(sample.counts) %in% names(population.counts))) {
## stop("all strata names in column '", data.stratum.column,
## "' must also appear in the loaded sampling frame.")
## }
## weights <- stratify.weights(pop_strata = population.counts,
## sample_strata = sample.counts)
## return(weights[df[[data.stratum.column]]])
## }
## <bytecode: 0x0000026b70132418>
## <environment: 0x0000026b70108cd8>## Warning in my_weigthing_function(main_dataset): 'weights' is used as a column
## name (will not be calculated from the sampling frame)## data.strata
## 5km_rural 5km_rural 20km_rural 20km_rural 20km_rural 20km_rural
## 0.2747648 0.2747648 0.6221156 0.6221156 0.6221156 0.6221156Note: A warning shows that if the column weights exists in the dataset, my_weigthing_function will not calculate weights. We need to remove the weights column from the previous example.
## data.strata
## 5km_rural 5km_rural 20km_rural 20km_rural 20km_rural 20km_rural
## 0.2747648 0.2747648 0.6221156 0.6221156 0.6221156 0.6221156To add the weights, we need to add a new column.
## [1] 0.2747648 0.2747648 0.6221156 0.6221156 0.6221156 0.6221156As for the previous example, we can check that only 1 weight is used per strata.
## # A tibble: 4 × 3
## # Groups: strata [4]
## strata my_weights n
## <chr> <table[1d]> <int>
## 1 20km_rural 0.6221156 407
## 2 20km_urban 1.6172529 404
## 3 5km_rural 0.2747648 402
## 4 5km_urban 1.4850825 404We can check that the sum of weights is equal to the number of interviews.
## [1] TRUE7.5 impactR, based on srvyr
impactR was at first designed for helping data officers to cover most of the research cycles’ parts, from producing cleaning log, cleaning data, and analyzing it. It is available for download here: https://gnoblet.github.io/impactR/.
The visualization (post-analysis) component has now been moved to package visualizeR: https://gnoblet.github.io/visualizeR/. Composite indicators now lies into humind : https://github.com/gnoblet/humind. Analysis is still a module of impactR; yet it will most probably shortly be moved to a smaller consolidated packages.
There are some vignettes/some documentation on the Github website. In particular this one for analysis: https://gnoblet.github.io/impactR/articles/analysis.html.
You can install and load it with:
##
## Attaching package: 'impactR'## The following objects are masked _by_ '.GlobalEnv':
##
## choices, cleaning_log7.5.1 Introduction
7.5.1.1 Caveats
Though it has been used for several research cycles, including MSNAs, there is no test designed in the package. One discrepancy will be corrected in the next version using function make_analysis_from_dap(): it for now assumes that all id_analysis are distinct and not NAs when using bind = TRUE. The next version will also have a new feature: unweighted counts and some automation for select one and select multiple questions.
Grouping is for now only possible for one variable. If you want to disagregate for instance by setting (rural vs. urban) and admin1, then you must mutate a new variable beforehand (it is as simple as pasting both columns).
7.5.1.2 Rational
The package is a wrapper around srvyr. The workflow is as follows:
- Prepare data: Get your data, weights, strata ready, and prepare a survey design object (using
srvyr::as_survey_design()). - Individual general analysis: It has a small family of functions
svy_*()such assvy_prop()which are wrappers around thesrvyr::survey_*()family in order to harmonize outputs. These functions can be used on their owns and covers mean, median, ration, proportion, interaction. - Individual analysis based on a Kobo tool: Prepare your Kobo tool, then use
make_analysis(). - Multiple analyses based on a Kobo tool: Prepare your Data Analysis Plan, then feed
make_analysis_from_dap().
7.5.1.3 Prepare data
Data must have been imported with ìmport_xlsx() or import_csv() or with janitor::clean_names(). This is to ensure that column names for multiple choices follow this pattern: “variable_choice1”, with an underscore between the variable name from the survey sheet and the choices from the choices sheet. For instance, for the main drinking water source (if multiple choice), it could be “w_water_source_surface_water” or “w_water_source_stand_pipe”.
7.5.2 First stage: prepare data and survey design
We start by downloading a dataset and the associated Kobo tool.
# The dataset (main and roster sheets), and the Kobo tool sheets are saved as a RDS object
all <- readRDS("inputs/REACH_HTI_dataset_MSNA-2022.RDS")
# Sheet main contains the HH data
main <- all$main
# Sheet survey contains the survey sheet
survey <- all$survey
# Sheet choices contains, well, the choices sheet
choices <- all$choicesLet’s prepare:
- data with
janitor::clean_names()to ensure lower case and only underscore (no dots, no /, etc.). - survey and choices: let’s separate column type from survey and rename the label column we want to keep to
label, otherwise most functions won’t work.
main <- main |>
# To get clean_names()
janitor::clean_names() |>
# To get better types
readr::type_convert()##
## ── Column specification ─────────────────────────────────────
## cols(
## .default = col_character()
## )
## ℹ Use `spec()` for the full column specifications.survey <- survey |>
# To get clean names
janitor::clean_names() |>
# To get two columns one for the variable type, and one for the list_name
impactR::split_survey(type) |>
# To get one column labelled "label" if multiple languages are used
dplyr::rename(label = label_francais)
choices <- choices |>
janitor::clean_names() |>
dplyr::rename(label = label_francais)Now that the dataset and the Kobo tool are loaded, we can prepare the survey design:
7.5.3 Second stage: One variable analysis
Let’s give a few examples for the svy_*() family.
# Median of respondent's age, with the confidence interval, no grouped analysis
impactR::svy_median(design, i_enquete_age, vartype = "ci")## # A tibble: 1 × 3
## median median_low median_upp
## <dbl> <dbl> <dbl>
## 1 42 42 43# Proportion of respondent's gender, with the confidence interval, by setting (rural vs. urban)
impactR::svy_prop(design, i_enquete_genre, milieu, vartype = "ci")## # A tibble: 4 × 5
## milieu i_enquete_genre prop prop_low prop_upp
## <chr> <chr> <dbl> <dbl> <dbl>
## 1 rural femme 0.598 0.570 0.626
## 2 rural homme 0.402 0.374 0.430
## 3 urbain femme 0.595 0.573 0.616
## 4 urbain homme 0.405 0.384 0.427# Ratio of the number of 0-17 yo children that attended school, with the confidence interval and no group
impactR::svy_ratio(design, e_formellet, c_total_age_3a_17a, vartype = "ci")## # A tibble: 1 × 3
## ratio ratio_low ratio_upp
## <dbl> <dbl> <dbl>
## 1 0.864 0.853 0.875# Top 6 common profiles between type of sanitation and source of drinking water, by setting
impactR::svy_interact(design, c(h_2_type_latrine, h_1_principal_source_eau), group = milieu, vartype = "ci") |> head(6)## # A tibble: 6 × 6
## milieu h_2_type_latrine h_1_principal_source_eau prop prop_low prop_upp
## <chr> <chr> <chr> <dbl> <dbl> <dbl>
## 1 rural trou_ouvert source_non_protege 0.122 0.104 0.140
## 2 rural defecation_air_libre source_non_protege 0.118 0.100 0.137
## 3 rural trou_ouvert robinet_public 0.0707 0.0564 0.0851
## 4 urbain toilette_chasse_eau reseau_dinepa 0.0679 0.0588 0.0770
## 5 urbain trou_ouvert robinet_public 0.0664 0.0543 0.0784
## 6 urbain toilette_chasse_eau camion_citerne 0.0526 0.0445 0.06077.5.4 Third stage: one analysis at a time with the Kobo tool
Now that we understands the svy_*() family of functions, we can use make_analysis() which is a wrapper around those for making analysis for medians, means, ratios, proportions of select ones and select multiples. The output of make_analysis() has standardized columns among all types of analysis and labels are pulled out if possible thanks to the survey and choices sheets.
Let’s say you don’t have a full DAP sheet, and you just want to make individual analyses.
1- Proportion for a single choice question:
# Single choice question, sanitation facility type
impactR::make_analysis(design, survey, choices, h_2_type_latrine, "prop_simple", level = 0.95, vartype = "ci")
# Same thing without labels, do not get labels of choices
impactR::make_analysis(design, survey, choices, h_2_type_latrine, "prop_simple", get_label = FALSE)
# Single choice question, by group setting (rural vs. urban) - group needs to be a character string here
impactR::make_analysis(design, survey, choices, h_2_type_latrine, "prop_simple", group = "milieu")2- Proportion overall: calculate the proportion with NAs as a category, this can be useful in case of a skip logic and you want to make the calculation over the whole population:
# Single choice question:
impactR::make_analysis(design, survey, choices, r_1_date_reception_assistance, "prop_simple_overall", none_label = "No assistance received, do not know, prefere not to say, or missing data")3- Multiple proportion:
# Multiple choice question, here computing self-reported priority needs
impactR::make_analysis(design, survey, choices, r_1_besoin_prioritaire, "prop_multiple", level = 0.95, vartype = "ci")
# Multiple choice question, with no label and with groups
impactR::make_analysis(design, survey, choices, r_1_besoin_prioritaire, "prop_multiple", get_label = FALSE, group = "milieu")4- Multiple proportion overall: calculate the proportion for each choice over the whole population (replaces NAs by 0s in the dummy columns):
# Multiple choice question, estimates calculated over the whole population.
impactR::make_analysis(design, survey, choices, r_1_besoin_prioritaire, "prop_multiple_overall")5- Mean, median and counting numeric as character:
# Mean of interviewee's age
impactR::make_analysis(design, survey, choices, i_enquete_age, "mean")
# Median of interviewee's age
impactR::make_analysis(design, survey, choices, i_enquete_age, "median")
# Proportion counting a numeric variable as a character variable
# Could be used for some particular numeric variables. For instance, below is the % of HHs by daily duration of access to electricity by setting (rural or urban).
impactR::make_analysis(design, survey, choices, a_6_acces_courant, "count_numeric", group = "milieu")6 - Last but not least, ratios (it does not consider NAs). For this, it is only necessary to write a character string with the two variable names separated by a comma:
# Let's calculate the % of children aged 3-17 that were registered to a formal school among the HHs that reported having children aged 3-17.
impactR::make_analysis(design, survey, choices, "e_formellet,c_total_age_3a_17a", "ratio")7.5.4.1 Fourth stage: Using a data analysis plan
Now we can use a dap, which mainly consists of a data frame with needed information for performing the analysis. One line is one requested analysis. All variables in the dap must exist in the dataset to be analysed. Usually an excel sheet is the easiest way to go as it can be co-constructed by both an AO and a DO.
# Here I produce a three-line DAP directly in R just for the sake of the example.
dap <- tibble::tibble(
# Column: unique analaysis id identifier
id_analysis = c("foodsec_1", "foodsec_3", "foodsec_3"),
# Column: sector/research question
rq = c("FSL", "FSL", "FSL"),
# Column: sub-sector, sub research question
sub_rq = c("Food security", "Food security", "Livelihoods"),
# Column: indicator name
indicator = c("% of HHs by FCS category", "% of HHs by HHS level", "% of HHs by LCSI category"),
# Column: recall period
recall = c("7 days", "30 days", "30 days"),
# Column: the variable name in the dataset
question_name = c("fcs_cat", "hhs_cat", "lcs_cat"),
# Column: subset (to be written by hand, does the calculation operates on a substed of the population)
subset = c(NA_character_, NA_character_, NA_character_),
# Column: analysis name to be passed to `make_analysis()`
analysis = c("prop_simple", "prop_simple", "prop_simple"),
# Column: analysis name used to be displayed later on
analysis_name = c("Proportion", "Proportion", "Proportion"),
# Column: if using analysis "prop_simple_overall", what is the label for NAs, passed through `make_analysis()`
none_label = c(NA_character_, NA_character_, NA_character_),
# Column: group label
group_name = "General population",
# Column: group variable from which to disaggregate (setting as an example)
group = c("milieu", "milieu", "milieu"),
# Column: level of confidence
level = c(0.95, 0.95, 0.95),
# Column: should NAs be removed?
na_rm = c("TRUE", "TRUE", "TRUE"),
# Column! var type, here we want confidence intervals
vartype = c("ci", "ci", "ci")
)
dap## # A tibble: 3 × 15
## id_analysis rq sub_rq indicator recall question_name subset analysis
## <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 foodsec_1 FSL Food security % of HHs… 7 days fcs_cat <NA> prop_si…
## 2 foodsec_3 FSL Food security % of HHs… 30 da… hhs_cat <NA> prop_si…
## 3 foodsec_3 FSL Livelihoods % of HHs… 30 da… lcs_cat <NA> prop_si…
## # … with 7 more variables: analysis_name <chr>, none_label <chr>,
## # group_name <chr>, group <chr>, level <dbl>, na_rm <chr>, vartype <chr>Let’s produce an analysis:
# Getting a list
an <- impactR::make_analysis_from_dap(design, survey, choices, dap)
# Note that if the variables' labels cannot be found in the Kobo tool, the variable's values are used as choices labels
# Getting a long dataframe
impactR::make_analysis_from_dap(design, survey, choices, dap, bind = TRUE) |> head(10)
# To perform the same analysis without grouping, you can just replace the "group" variable by NAs
dap |>
srvyr::mutate(group = rep(NA_character_, 3)) |>
impactR::make_analysis_from_dap(design, survey, choices, dap = _, bind = TRUE) |>
head(10)7.6 EXAMPLE :: Survey analysis using illuminate
illuminate is designed for making the data analysis easy and less time consuming. The package is based on tidyr, dplyr,srvyr packages.You can install the package from here. HOWEVER the package is not maintained by the author anymore and it doesnt includes any tests.
7.6.2 Step 1:: Read data
Read data with read_sheets(). This will make sure your data is stored as appropriate data type. It is important to make sure that all the select multiple are stored as logical, otherwise un weighted count will be wrong. It is recommended to use the read_sheets() to read the data. use `?read_sheets() for more details.
# [recommended] read_sheets("[datapath]",data_type_fix = T,remove_all_NA_col = T,na_strings = c("NA",""," "))
data_up<-read.csv("inputs/UKR2007_MSNA20_HH_dataset_main_rcop.csv")The above code will give a dataframe called data_up
7.6.3 Step OPTIONAL:: Preaparing the data
ONLY APPLICABLE WHEN YOU ARE NOT READING YOUR DATA WITH read_sheets()
7.6.4 Step 2:: Weight calculation
To do the weighted analysis, you will need to calculate the weights. If your dataset already have the weights column then you can ignore this part
7.6.5 Read sampleframe
This will give a data frame called sampling_frame
# dummy code.
weights <- data_up %>% group_by(governorate1) %>% summarise(
survey_count = n()
) %>% left_join(sampling_frame,by =c("governorate1"="strata.names")) %>%
mutate(sample_global=sum(survey_count),
pop_global=sum(population),
survey_weight= (population/pop_global)/(survey_count/sample_global)) %>% select(governorate1,survey_weight)7.6.7 Step 3.1:: Weighted analysis
To do the weighted analysis, you need to set weights parameter to TRUE and provide weight_column and strata. You should define the name of the data set weight column in the weight_column parameter and name of the strata column name from the data set.
Additionally, please define the name of the columns which you would like to analyze in vars_to_analyze. Default will analyze all the variables that exist in the data set.
Lastly, please define the sm_sep parameter carefully. It must be either / or ,. Use / when your select multiple type questions are separated by / (example- parent_name/child_name1,parent_name/child_name2,
parent_name/child_name3…..). On the other hand use . when the multiple choices are separated by .. (example - parent_name.child_name1,parent_name.child_name2,parent_name.child_name3)
7.6.8 Step 3.2:: Unweighted analysis
To do unweighted analysis, you need to set the weights parameter FALSE.
columns_to_analyze <- data_up[20:50] %>% names() ## Defining names of the variables to be analyzed.
overall_analysis <- survey_analysis(df = data_up,weights = F,vars_to_analyze = columns_to_analyze )## [1] "b10_1_hohh_vulnerability_other"
## [1] "b11_hohh_chronic_illness.blood_pressure_diseases"
## [1] "b11_hohh_chronic_illness.cardiovascular_disease"
## [1] "b11_hohh_chronic_illness.diabetes_need_insulin"
## [1] "b11_hohh_chronic_illness.diabetes_does_not_need_insulin"
## [1] "b11_hohh_chronic_illness.chronic_respiratory_condition"
## [1] "b11_hohh_chronic_illness.musculoskeletal_system_and_joints"
## [1] "b11_hohh_chronic_illness.cancer"
## [1] "b11_hohh_chronic_illness.neurological"
## [1] "b11_hohh_chronic_illness.sensory_disorder"
## [1] "b11_hohh_chronic_illness.gastrointestinal_digestive_tract_incl_liver_gallbladder_pancreas_diseases"
## [1] "b11_hohh_chronic_illness.genitourinary_system_diseases"
## [1] "b11_hohh_chronic_illness.endocrine_system_thyroid_gland_and_other_diseases"
## [1] "b11_hohh_chronic_illness.other_specify"
## [1] "b11_1_hohh_chronic_illness_other"
## [1] "b12_situation_description"
## [1] "b12_1_situation_description_other"
## [1] "b13_why_hohh_unemployed"
## [1] "b13_1_why_hohh_unemployed_other"
## [1] "b14_sector_hohh_employed"
## [1] "b14_1_sector_hohh_employed_other"
## [1] "b15_hohh_income"
## [1] "b16_hohh_pension_eligible"
## [1] "b17_hohh_pension_receive"
## [1] "b18_hohh_benefit_eligible"
## [1] "b19_hohh_benefit_receive"
## [1] "b20_hohh_level_education"
## [1] "hohh_age_18"
## [1] "age_18"
## [1] "b21_num_additional_hh_members"
## [1] "b11_hohh_chronic_illness.blood_pressure_diseases"
## [1] "b11_hohh_chronic_illness.cardiovascular_disease"
## [1] "b11_hohh_chronic_illness.diabetes_need_insulin"
## [1] "b11_hohh_chronic_illness.diabetes_does_not_need_insulin"
## [1] "b11_hohh_chronic_illness.chronic_respiratory_condition"
## [1] "b11_hohh_chronic_illness.musculoskeletal_system_and_joints"
## [1] "b11_hohh_chronic_illness.cancer"
## [1] "b11_hohh_chronic_illness.neurological"
## [1] "b11_hohh_chronic_illness.sensory_disorder"
## [1] "b11_hohh_chronic_illness.gastrointestinal_digestive_tract_incl_liver_gallbladder_pancreas_diseases"
## [1] "b11_hohh_chronic_illness.genitourinary_system_diseases"
## [1] "b11_hohh_chronic_illness.endocrine_system_thyroid_gland_and_other_diseases"
## [1] "b11_hohh_chronic_illness.other_specify"7.6.9 Step 3.3:: Weighted and disaggregated by gender analysis
Use ?survey_analysis() to know about the perameters.
7.8 Analysis - top X / ranking (select one and select multiple)
First: select_one question
The indicator of interest is e1_shelter_type and the options are:
- solid_finished_house
- solid_finished_apartment
- unfinished_nonenclosed_building
- collective_shelter
- tent
- makeshift_shelter
- none_sleeping_in_open
- other_specify
- dont_know_refuse_to_answer
# Top X/ranking for select_one type questions
select_one_topX <- function(df, question_name, X=3, weights_col=NULL, disaggregation_col=NULL){
# test message
if(length(colnames(df)[grepl(question_name, colnames(df), fixed = T)]) == 0) {
stop(print(paste("question name:", question_name, "doesn't exist in the main dataset. Please double check and make required changes!")))
}
#### no disag
if(is.null(disaggregation_col)){
#no weights
if(is.null(weights_col)){
df <- df %>%
filter(!is.na(!!sym(question_name))) %>%
group_by(!!sym(question_name)) %>%
summarize(n=n()) %>%
mutate(percentages = round(n / sum(n) * 100, digits = 2)) %>%
select(!!sym(question_name), n, percentages) %>%
arrange(desc(percentages)) %>%
head(X)
}
# weights
if(!is.null(weights_col)){
df <- df %>%
filter(!is.na(!!sym(question_name))) %>%
group_by(!!sym(question_name)) %>%
summarize(n=sum(!!sym(weights_col))) %>%
mutate(percentages = round(n / sum(n) * 100, digits = 2)) %>%
select(!!sym(question_name), n, percentages) %>%
arrange(desc(percentages)) %>%
head(X)
}
}
#### with disag
if(!is.null(disaggregation_col)){
#no weights
if(is.null(weights_col)){
df <- df %>%
filter(!is.na(!!sym(question_name))) %>%
group_by(!!sym(disaggregation_col), !!sym(question_name)) %>%
summarize(n=n()) %>%
mutate(percentages = round(n / sum(n) * 100, digits = 2)) %>%
select(!!sym(disaggregation_col), !!sym(question_name), n, percentages)
df <- split(df, df[[disaggregation_col]])
df <- lapply(df, function(x){
x %>% arrange(desc(percentages)) %>%
head(X)
})
}
## weights
if(!is.null(weights_col)){
df <- df %>%
filter(!is.na(!!sym(question_name))) %>%
group_by(!!sym(disaggregation_col), !!sym(question_name)) %>%
summarize(n=sum(!!sym(weights_col))) %>%
mutate(percentages = round(n / sum(n) * 100, digits = 2)) %>%
select(!!sym(disaggregation_col), !!sym(question_name), n, percentages)
df <- split(df, df[[disaggregation_col]])
df <- lapply(df, function(x){
x %>% arrange(desc(percentages)) %>%
head(X)
})
}
}
return(df)
}
# return top 4 shelter types
shelter_type_top4 <- select_one_topX(df = main_dataset, question_name = "e1_shelter_type", X = 4)Second: select_multiple question
The indicator of interest is e3_shelter_enclosure_issues and the options are:
- no_issues
- lack_of_insulation_from_cold
- leaks_during_light_rain_snow
- leaks_during_heavy_rain_snow
- limited_ventilation_less_than_05m2_ventilation_in_each_room_including_kitchen
- presence_of_dirt_or_debris_removable
- presence_of_dirt_or_debris_nonremovable
- none_of_the_above
- other_specify
- dont_know
# Top X/ranking for select_multiple type questions
select_multiple_topX <- function(df, question_name, X = 3, separator=".", weights_col=NULL, disaggregation_col=NULL) {
# test message
if(length(colnames(df)[grepl(question_name, colnames(df), fixed = T)]) == 0){
stop(print(paste("question name:", question_name, "doesn't exist in the main dataset. Please double check and make required changes!")))
}
#work on the select multiple questions
df_mult <- df %>%
select(colnames(df)[grepl(paste0(question_name, separator), colnames(df), fixed = T)]) %>%
mutate_all(as.numeric)
# Keeping only the options names in the colnames
colnames(df_mult) <- gsub(paste0(question_name, separator), "", colnames(df_mult))
# if question was not answered
if(all(is.na(df_mult))) warning("None of the choices was selected!")
#if weights, multiply each dummy by the weight of the HH
if(!is.null(weights_col)){
df_weight <- dplyr::pull(df, weights_col)
df_mult <- df_mult %>% mutate(across(everything(), ~. *df_weight))
}
### calculate top X percentages
# no disag
if(is.null(disaggregation_col)){
df <- df_mult %>%
pivot_longer(cols = everything(), names_to = question_name, values_to = "choices") %>%
group_by(!!sym(question_name), .drop=F) %>%
summarise(n = sum(choices, na.rm = T),
percentages = round(sum(choices, na.rm = T) / n() * 100, digits = 2)) %>% # remove NA values from percentages calculations
arrange(desc(percentages)) %>%
head(X) # keep top X percentages only
}
# disag
if(!is.null(disaggregation_col)){
df <- df_mult %>%
bind_cols(df %>% select(disaggregation_col))
df <- split(df, df[[disaggregation_col]]) #split
df <- map(df, function(x){
x %>% select(-disaggregation_col) %>%
pivot_longer(cols = everything(), names_to = question_name, values_to = "choices") %>%
group_by(!!sym(question_name)) %>%
summarise(n = sum(choices, na.rm = T),
percentages = round(sum(choices, na.rm = T) / n() * 100, digits = 2)) %>% # remove NA values from percentages calculations
select(!!sym(question_name), n, percentages) %>%
arrange(desc(percentages)) %>%
head(X) # keep top X percentages only
})
}
return(df)
}
# return top 7 shelter enclosure issues
shelter_enclosure_issues_top7 <- select_multiple_topX(df = main_dataset, question_name = "e3_shelter_enclosure_issues", X = 7)7.10 Hypothesis testing
7.10.1 T-test
7.10.1.1 What is a T-test
A t-test is a type of inferential statistic used to determine if there is a significant difference between the means of two groups. A t-test is used as a hypothesis testing tool, which allows testing of an assumption applicable to a population.
7.10.1.2 How it works
Mathematically, the t-test takes a sample from each of the two sets and establishes the problem statement by assuming a null hypothesis that the two means are equal. Based on the applicable formulas, certain values are calculated and compared against the standard values, and the assumed null hypothesis is accepted or rejected accordingly.
If the null hypothesis qualifies to be rejected, it indicates that data readings are strong and are probably not due to chance.
7.10.1.3 T-Test Assumptions
The first assumption made regarding t-tests concerns the scale of measurement. The assumption for a t-test is that the scale of measurement applied to the data collected follows a continuous or ordinal scale, such as the scores for an IQ test.
The second assumption made is that of a simple random sample, that the data is collected from a representative, randomly selected portion of the total population.
The third assumption is the data, when plotted, results in a normal distribution, bell-shaped distribution curve.
The final assumption is the homogeneity of variance. Homogeneous, or equal, variance exists when the standard deviations of samples are approximately equal.
7.10.1.4 Example
We are going to look at the income of the household for female/male headed household. The main_dataset doesn’t contain the head of household gender information, so we are creating and randomly populating the gender variable.
set.seed(10)
main_dataset$hoh_gender = sample(c('Male', 'Female'), nrow(main_dataset), replace=TRUE)
main_dataset$b15_hohh_income = as.numeric(main_dataset$b15_hohh_income)##
## Welch Two Sample t-test
##
## data: b15_hohh_income by hoh_gender
## t = 0.19105, df = 1525.6, p-value = 0.8485
## alternative hypothesis: true difference in means between group Female and group Male is not equal to 0
## 95 percent confidence interval:
## -270.3420 328.6883
## sample estimates:
## mean in group Female mean in group Male
## 3459.044 3429.871In the result above :
t is the t-test statistic value (t = -0.19105), df is the degrees of freedom (df= 1525.6), p-value is the significance level of the t-test (p-value = 0.8485). conf.int is the confidence interval of the means difference at 95% (conf.int = [-270.3420, 328.6883]); sample estimates is the mean value of the sample (mean = 3459.044, 3429.871).
Interpretation:
The p-value of the test is 0.8485, which is higher than the significance level alpha = 0.05. We can’t reject the null hypothesis and we can’t conclude that men headed household average income is significantly different from women headed household average income. Which makes sense in this case given that the gender variable was randomly generated.
7.10.2 ANOVA
7.10.2.1 What is ANOVA
Similar to T-test, ANOVA is a statistical test for estimating how a quantitative dependent variable changes according to the levels of one or more categorical independent variables. ANOVA tests whether there is a difference in means of the groups at each level of the independent variable.
The null hypothesis (H0) of the ANOVA is no difference in means, and the alternate hypothesis (Ha) is that the means are different from one another.
The T-test is used to compare the means between two groups, whereas ANOVA is used to compare the means among three or more groups.
7.10.2.2 Types of ANOVA
There are two main types of ANOVA: one-way (or unidirectional) and two-way. A two-way ANOVA is an extension of the one-way ANOVA. With a one-way, you have one independent variable affecting a dependent variable. With a two-way ANOVA, there are two independents. For example, a two-way ANOVA allows a company to compare worker productivity based on two independent variables, such as salary and skill set. It is utilized to observe the interaction between the two factors and tests the effect of two factors at the same time.
7.10.2.3 Example
We are using the same example as the previous test.
## Df Sum Sq Mean Sq F value Pr(>F)
## hoh_gender 1 3.291e+05 329075 0.036 0.849
## Residuals 1546 1.408e+10 9109185
## 69 observations deleted due to missingnessAs the p-value is more than the significance level 0.05, we can’t conclude that there are significant differences between the hoh_gender groups.
7.10.2.4 Bonus : Visualization of the data to confirm the findinds
##
## Attaching package: 'ggpubr'## The following objects are masked from 'package:spatstat.geom':
##
## border, rotate## The following object is masked from 'package:expss':
##
## compare_meansggline(main_dataset, x = "hoh_gender", y = "b15_hohh_income",
add = c("mean_se", "jitter"),
order = c("Female", "Male"),
ylab = "HH income", xlab = "hoh gender")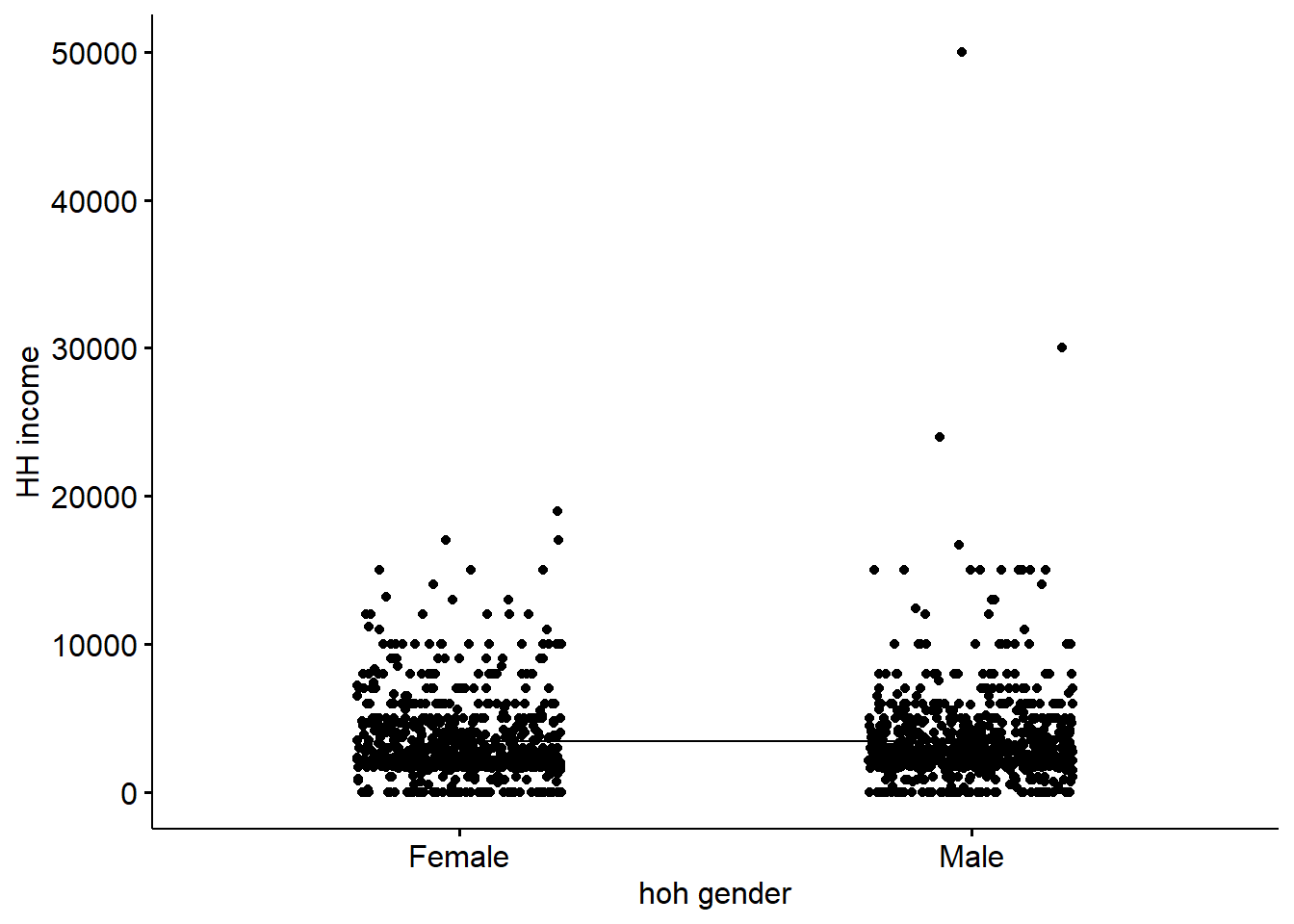
7.10.3 chi-squares
Pearson`s chi-squared test. Chi-squared test is a statistical hypothesis test, based on comparing expected and observed frequencies of 2-way distribution (2-way table).
There are three types of tests (hypothesis) that could be tested with chi-squared: independence of variables, homogenity of distribution of the characteristic across “stratas” and goodness of fit (whether the actual distribution is different from the hypothetical one). Two former versions use the same calculation.
Chi-squared test is a non-parametric test. It is used with nominal to nominal (nominal to ordinal) scales. For running chi-squared test, each cell of the crosstab should contain at least 5 observations.
Note, that chi-squared test indicates significance of differences in values on a table level. Thus, it is impossible to track significance in differences for particular options.
(For example, it is possible to say that satisfaction with public transport and living in urban / rural areas are connected. However, we can conclude from the test itself, that “satisfied” and “dissatisfied” options differ, while “indifferent” does not. By the way, it is possible to come up with this conclusion while interpreting a corresponding crosstab. Also, chi-squared test can tell nothing about the strength and the direction of a liaison between variables).
7.10.3.1 Chi-squared test for independence
The test for weighted data is being run with the survey library
The use of the survey package functions requires specifying survey design object (which reslects sampling approach).
main_dataset <- read.csv("inputs/UKR2007_MSNA20_HH_dataset_main_rcop.csv", na.strings = "", stringsAsFactors = F)
svy_design <- svydesign(data=main_dataset,
id = ~1,
strata = main_dataset$strata,
probs = NULL,
weights = main_dataset$stratum.weight)Specifying function for getting chi-squared outputs.
Hereby test Chi^2 test of independence is used. So, the test is performed to find out if strata variable and target variable are independent, H0 - the distribution of target variable values is equal across all stratas H1 - strata and target var are not independent, so distribution varies across stratas.
Apart from getting test statistic and p value, we would like to detect manually what are the biggest and the smallest values across groups/stratas (in case if the difference is significant and it makes sense to do it).
Hereby test Chi^2 test of independence is used. So, the test is performed to find out if strata variable and target variable are independent, H0 - the distribution of target var. values is equal across all stratas H1 - strata and target var are not independent, so distribution varies across stratas.
Within the Survey package, the suggested function uses Scott & Rao adjustment for complex survey design
#####Simple single call of the test.
Let’s say that the research aim is to understand whether the main water source is dependent on the area where HH lives (strata variable).
Thus, in our case: H0 - the distribution of water source is equal across stratas and these variables are independent H1 - the distribution of water source differs by strata and these variables are not independent.
Running the chi-squared test:
(Survey version - requires a survey design object)
##
## Pearson's X^2: Rao & Scott adjustment
##
## data: svychisq(~f11_dinking_water_source + strata, design = svy_design, statistic = "Chisq")
## X-squared = 244.62, df = 21, p-value < 2.2e-16The current output presents the result of Pearson’s chi-squared test (Rao & Scott adjustment accounts for weighted data) [https://www.researchgate.net/publication/38360147_On_Simple_Adjustments_to_Chi-Square_Tests_with_Sample_Survey_Data].
The output consist of three parameters: 1. X-squared - observed value of chi-squared parameter, 2. df - “degrees of freedom” or number of independent categories in data -1 Having only these two elements it is possible to see, if with the current number of df, the value of chi-squared parameter is greater than the critical value for the chosen level of probability, using (tables of chi-squared critical values) [https://www.itl.nist.gov/div898/handbook/eda/section3/eda3674.htm] Fortunately, R provides us with the third parameter, which allows us to skip this step. 3. p-value - stands for the probability of getting value of the chi-squared parameter greater or equal to the given one assuming that H0 is true. If p-value < 0.05 (for 95% confidence level, or 0.01 for 99%), the H0 can be rejected.
In the current example, as p-value is very low, H0 can be rejected, and hence, strata variable and drinking water source are not independent.
(Weights package - can work with the dataframe directly, without the survey design object)
wtd.chi.sq(var1 = main_dataset$f11_dinking_water_source, var2 = main_dataset$strata, weight = main_dataset$stratum.weight)## Chisq df p.value
## 2.446205e+02 2.100000e+01 4.928158e-40Specifying the function for crosstabs with chi-squared tests for independence.
#####Functions to have test results along with crosstabs
- The function with survey design object.
Apart from getting test statistic and p value, we would like to detect manually what are the biggest and the smallest values across groups/stratas (in case if the difference is significant and it makes sense to do it).
The function also helps to find out smallest and the largest values per row
chi2_ind_extr <- function(x, y, design){
my_var <- y
# running Pearson's chi-squared test
s_chi <- svychisq(as.formula(paste0("~", x,"+", y)), design, statistic = "Chisq")
# extracting basic parameters
wmy_t <- as.data.frame(s_chi$observed)
wmy_t$p <- s_chi$p.value
wmy_t$chi_adj <- s_chi$statistic
wmy_t$df <- s_chi$parameter
#indicating significance
wmy_t$independence <- ifelse(wmy_t$p < 0.05, "h1", "h0")
colnames(wmy_t)[1:2] <- c("var_x", "var_y")
wmy_t_w <- spread(wmy_t, var_y, Freq)
wmy_t_w[,1] <- paste0(x, " ", wmy_t_w[,1])
colnames(wmy_t_w)[1] <- "variable"
# getting percent from weighted frequencies
cols_no_prop <- c("variable", "p", "independence", "max", "min", "chi_adj", "df")
wmy_t_w[, !colnames(wmy_t_w) %in% cols_no_prop] <- lapply(wmy_t_w[, !colnames(wmy_t_w) %in% cols_no_prop], function(x){x/sum(x, na.rm = T)})
# indicating extremum values
cols_to_compare <- colnames(wmy_t_w)[!colnames(wmy_t_w) %in% c("p", "chi_adj", "df", "independence", "min", "max", "variable")]
wmy_t_w$max <- ifelse(wmy_t_w$independence == "h1", cols_to_compare[apply(wmy_t_w, 1, which.max)], 0)
wmy_t_w$min <- ifelse(wmy_t_w$independence == "h1", cols_to_compare[apply(wmy_t_w, 1, which.min)], 0)
#adding Overall value to the table
ovl_tab <- as.data.frame(prop.table(svytable(as.formula(paste0("~",x)), design)))
colnames(ovl_tab) <- c("options", "overall")
wmy_t_w <- wmy_t_w %>% bind_cols(ovl_tab)
return(wmy_t_w)
}The use with a single-choice question
## variable
## 1 f11_dinking_water_source bottled_water_water_purchased_in_bottles
## 2 f11_dinking_water_source drinking_water_from_water_kiosk_booth_with_water_for_bottling
## 3 f11_dinking_water_source other_specify
## 4 f11_dinking_water_source personal_well
## 5 f11_dinking_water_source public_well_or_boreholes_shared_access
## 6 f11_dinking_water_source tap_drinking_water_centralized_water_supply
## 7 f11_dinking_water_source technical_piped_water
## 8 f11_dinking_water_source trucked_in_water_truck_with_a_tank_etc
## p chi_adj df independence 20km_rural 20km_urban 5km_rural
## 1 9.386389e-50 244.6205 21 h1 0.02457002 0.07673267 0.034825871
## 2 9.386389e-50 244.6205 21 h1 0.19901720 0.29950495 0.131840796
## 3 9.386389e-50 244.6205 21 h1 0.01228501 0.00000000 0.007462687
## 4 9.386389e-50 244.6205 21 h1 0.32432432 0.09653465 0.482587065
## 5 9.386389e-50 244.6205 21 h1 0.15724816 0.05445545 0.151741294
## 6 9.386389e-50 244.6205 21 h1 0.22358722 0.43316832 0.144278607
## 7 9.386389e-50 244.6205 21 h1 0.00000000 0.00000000 0.000000000
## 8 9.386389e-50 244.6205 21 h1 0.05896806 0.03960396 0.047263682
## 5km_urban max min
## 1 0.032178218 5km_rural 20km_urban
## 2 0.274752475 5km_rural 20km_urban
## 3 0.007425743 5km_rural <NA>
## 4 0.121287129 5km_rural 20km_urban
## 5 0.099009901 5km_rural 20km_urban
## 6 0.383663366 5km_rural 20km_urban
## 7 0.004950495 5km_rural <NA>
## 8 0.076732673 5km_rural 20km_urban
## options overall
## 1 bottled_water_water_purchased_in_bottles 0.049170548
## 2 drinking_water_from_water_kiosk_booth_with_water_for_bottling 0.263132750
## 3 other_specify 0.005188695
## 4 personal_well 0.167758525
## 5 public_well_or_boreholes_shared_access 0.093728457
## 6 tap_drinking_water_centralized_water_supply 0.362248561
## 7 technical_piped_water 0.001836837
## 8 trucked_in_water_truck_with_a_tank_etc 0.056935627In the given output, p indicates chi-squared test p-value, chi_adj - value of chi-squared parameter, df - degrees of freedom, independence - indicates which hypothesis is accepted, h0 or h1. In the current function, H1 is accepted if p < 0.05. In case H0 is rejected and H1 accepted, in min and max columns minimum and maximum y variable categories are given.
The use with a multiple-choice question.
The test would not work if any of dummies consists from zeros only, it need to be checked in advance, thus, let’s filter such variables out from the vector of variable names
Getting full range of dummies from the question
Defining a function which would remove zero-sum variables for us
if the absence of empty cells for each strata is needed, per_cell = T
non_zero_for_test <- function(var, strata, dataset, per_cell = F){
sum <- as.data.frame(sapply(dataset[, names(dataset) %in% var], sum))
sum <- cbind(rownames(sum), sum)
filtered <- sum[sum[,2] > 0,1]
tab <- as.data.frame(sapply(dataset[,names(dataset) %in% filtered], table, dataset[, strata]))
ntab <- names(tab)
#if the absence of empty cells for each strata is needed, per_cell = T
tab <- tab[, which(colnames(tab) %in% ntab)]
zeros <- apply(apply(tab, 2, function(x) x==0), 2, any)
zeros <- zeros[zeros == F]
zeros <- as.data.frame(zeros)
zeros <- cbind(rownames(zeros), zeros)
zeros_lab <- as.vector(unlist(strsplit(zeros[,1], split = ".Freq")))
result <- c()
if(per_cell == T){
result <- zeros_lab
} else {
result <- ntab
}
message(paste0(abs(length(var) - length(result)), " variables were removed because of containing zeros in stratas"))
return(result)
}Using the test with multiple choice question.
## 2 variables were removed because of containing zeros in stratasf12_sig <- lapply(nz_f12, chi2_ind_extr, y = "strata", design = svy_design)
f12_sig_df <- Reduce(rbind, f12_sig)
#removing 0 ("not selected") options
f12_sig_df <- f12_sig_df %>% filter(options == 1)
f12_sig_df## variable
## 1 f12_drinking_water_treat.cleaning_with_chemicals_chlorination 1
## 2 f12_drinking_water_treat.water_precipitation 1
## 3 f12_drinking_water_treat.filtering_the_water_pitcher_filter 1
## 4 f12_drinking_water_treat.filtering_the_water_reverse_osmosis_filter 1
## 5 f12_drinking_water_treat.boiling 1
## 6 f12_drinking_water_treat.percolation 1
## 7 f12_drinking_water_treat.other_specify 1
## 8 f12_drinking_water_treat.do_not_process_purify 1
## p chi_adj df independence 20km_rural 20km_urban 5km_rural
## 1 0.03100696 4.780006 3 h1 0.000000000 0.004950495 0.000000000
## 2 0.02775288 7.977278 3 h1 0.073710074 0.126237624 0.104477612
## 3 0.55635016 1.828231 3 h0 0.046683047 0.069306931 0.059701493
## 4 0.44029529 2.347539 3 h0 0.027027027 0.037128713 0.017412935
## 5 0.02724163 8.191084 3 h1 0.260442260 0.274752475 0.223880597
## 6 0.24801653 3.459406 3 h0 0.004914005 0.002475248 0.007462687
## 7 0.28029092 2.860754 3 h0 0.004914005 0.002475248 0.000000000
## 8 0.01762880 9.011919 3 h1 0.624078624 0.551980198 0.674129353
## 5km_urban max min options overall
## 1 0.00000000 5km_rural <NA> 1 0.002000313
## 2 0.14108911 5km_rural 20km_urban 1 0.122036605
## 3 0.06930693 0 0 1 0.065108190
## 4 0.04207921 0 0 1 0.036036995
## 5 0.20792079 5km_rural 20km_urban 1 0.244239386
## 6 0.00000000 0 0 1 0.002279393
## 7 0.00000000 0 0 1 0.001769625
## 8 0.60643564 5km_rural 20km_urban 1 0.591818943- The function without the survey design object
chi2_ind_wtd <- function(x, y, weight, dataset){
i <- 1
table_new <- data.frame()
for (i in 1:length(x)) {
ni <- x[i]
ti <- dataset %>%
filter(!is.na(!!sym(y))) %>%
group_by(!!sym(y), !!sym(ni)) %>%
filter(!is.na(!!sym(ni))) %>%
summarise(wtn = sum(!!sym(weight)), n = n()) %>%
mutate(prop = wtn/sum(wtn), varnam = ni)
names(ti) <- c("var_ind", "var_dep_val", "wtn", "n", "prop", "dep_var_nam")
ti <- ti %>% ungroup() %>% mutate(base = sum(c_across(n)))
chi_test <- wtd.chi.sq(var1 = dataset[, paste0(y)], var2 = dataset[, paste0(ni)], weight = dataset[, paste0(weight)])
ti$chisq <- chi_test[1]
ti$df <- chi_test[2]
ti$p <- chi_test[3]
ti$hypo <- ifelse(ti$p < 0.05, "h1", "h0")
table_new <- rbind(table_new, ti)
}
table_cl_new_1 <- table_new %>%
#filter(var_dep_val != "0") %>%
filter(!is.na(var_dep_val))
#mutate(var_id = paste0(dep_var_nam, ".", var_dep_val))
table_cl_new_1 <- table_cl_new_1[, -c(3, 4)]
table_sprd <- tidyr::spread(table_cl_new_1, var_ind, prop) %>%
arrange(dep_var_nam)
return(table_sprd)
}The use with the single choice question
## `summarise()` has grouped output by 'strata'. You can
## override using the `.groups` argument.## # A tibble: 8 × 11
## var_dep_val dep_var_nam base chisq df p hypo `20km_rural`
## <chr> <chr> <int> <dbl> <dbl> <dbl> <chr> <dbl>
## 1 bottled_water_water… f11_dinkin… 1617 245. 21 4.93e-40 h1 0.0246
## 2 drinking_water_from… f11_dinkin… 1617 245. 21 4.93e-40 h1 0.199
## 3 other_specify f11_dinkin… 1617 245. 21 4.93e-40 h1 0.0123
## 4 personal_well f11_dinkin… 1617 245. 21 4.93e-40 h1 0.324
## 5 public_well_or_bore… f11_dinkin… 1617 245. 21 4.93e-40 h1 0.157
## 6 tap_drinking_water_… f11_dinkin… 1617 245. 21 4.93e-40 h1 0.224
## 7 technical_piped_wat… f11_dinkin… 1617 245. 21 4.93e-40 h1 NA
## 8 trucked_in_water_tr… f11_dinkin… 1617 245. 21 4.93e-40 h1 0.0590
## # … with 3 more variables: `20km_urban` <dbl>, `5km_rural` <dbl>,
## # `5km_urban` <dbl>The use with multiple choice questions
names <- names(main_dataset)
f12_names <- names[grepl("f12_drinking_water_treat.", names)]
f12_sig2 <- lapply(f12_names, chi2_ind_wtd, y = "strata", weight = "stratum.weight", main_dataset)
f12_tab_df <- Reduce(rbind, f12_sig2)
f12_tab_df## # A tibble: 18 × 11
## var_dep_val dep_var_nam base chisq df p hypo `20km_rural`
## <int> <chr> <int> <dbl> <dbl> <dbl> <chr> <dbl>
## 1 0 f12_drinking_water_t… 1617 4.78 3 0.189 h0 1
## 2 1 f12_drinking_water_t… 1617 4.78 3 0.189 h0 NA
## 3 0 f12_drinking_water_t… 1617 7.98 3 0.0465 h1 0.926
## 4 1 f12_drinking_water_t… 1617 7.98 3 0.0465 h1 0.0737
## 5 0 f12_drinking_water_t… 1617 1.83 3 0.609 h0 0.953
## 6 1 f12_drinking_water_t… 1617 1.83 3 0.609 h0 0.0467
## 7 0 f12_drinking_water_t… 1617 2.35 3 0.503 h0 0.973
## 8 1 f12_drinking_water_t… 1617 2.35 3 0.503 h0 0.0270
## 9 0 f12_drinking_water_t… 1617 8.19 3 0.0422 h1 0.740
## 10 1 f12_drinking_water_t… 1617 8.19 3 0.0422 h1 0.260
## 11 0 f12_drinking_water_t… 1617 3.46 3 0.326 h0 0.995
## 12 1 f12_drinking_water_t… 1617 3.46 3 0.326 h0 0.00491
## 13 0 f12_drinking_water_t… 1617 2.86 3 0.414 h0 0.995
## 14 1 f12_drinking_water_t… 1617 2.86 3 0.414 h0 0.00491
## 15 0 f12_drinking_water_t… 1617 0 0 1 h0 1
## 16 0 f12_drinking_water_t… 1617 0 0 1 h0 1
## 17 0 f12_drinking_water_t… 1617 9.01 3 0.0291 h1 0.376
## 18 1 f12_drinking_water_t… 1617 9.01 3 0.0291 h1 0.624
## # … with 3 more variables: `20km_urban` <dbl>, `5km_rural` <dbl>,
## # `5km_urban` <dbl>7.10.4 Chi-squared Goodness of Fit test
Goodness of test chi-squared test is used to compare the observed distribution with the hypothetical one,
For instance, we would like to test the hypothesis, that 90% of HHs had soap in their HHs at the moment of the survey.
main_dataset <- main_dataset %>% mutate(soap = case_when(
p16_soap_household == "dont_know_refuse_to_answer" ~ "NA",
p16_soap_household == "no" ~ "no",
p16_soap_household == "yes_soap_is_not_shown" ~ "yes",
p16_soap_household == "yes_soap_is_shown" ~ "yes"
))
soap_prob <- as.data.frame(prop.table(table(main_dataset$soap)))
soap_prob <- soap_prob[, 2]
# please, note that this is a function call with unweighted data!
chisq.test(soap_prob, p = c(0, 0.1, 0.9))## Warning in chisq.test(soap_prob, p = c(0, 0.1, 0.9)): Chi-squared approximation
## may be incorrect##
## Chi-squared test for given probabilities
##
## data: soap_prob
## X-squared = Inf, df = 2, p-value < 2.2e-16As p value is lower than the chosen confidence level, hypothesis of equality of the current distribution to hypothetical 90% / 10% of soap-owners can be rejected.
Defining a function for checking the distribution within a particular group, against the “hypothetical” overall sample.
For the overall sample we would need a variable which has the same value across all observations (e.g. informed consent == yes etc.)
chi2_GoF <- function(name, stratum_hyp, stratum_2, value_stratum_hyp, value_stratum_2, data, weights){
i <- 1
table_1 <- data.frame()
for (i in 1:length(name)) {
ni <- name[i]
ti <- data %>%
group_by(!!sym(ni)) %>% filter(!!sym(stratum_hyp) == paste0(value_stratum_hyp)) %>%
filter(!is.na(!!sym(ni))) %>%
summarise(wtn = sum(!!sym(weights)), n = n()) %>%
mutate(prop = wtn/sum(wtn), base = sum(wtn), cnt = sum(n))%>%
mutate(varnam = ni)
names(ti) <- c("var", "wtn", "n", "prop", "base_w", "count", "nam")
table_1 <- rbind(table_1, ti)
}
table_cl_1 <- table_1 %>% filter(!is.na(var)) %>%
mutate(var_id = paste0(nam, ".", var))
i <- 1
table_2 <- data.frame()
for (i in 1:length(name)) {
ni <- name[i]
ti <- data %>%
group_by(!!sym(ni)) %>% filter(!!sym(stratum_2) == paste0(value_stratum_2)) %>%
filter(!is.na(!!sym(ni))) %>%
summarise(wtn = sum(!!sym(weights)), n = n()) %>%
mutate(prop = wtn/sum(wtn), base = sum(wtn), cnt = sum(n))%>%
mutate(varnam = ni)
names(ti) <- c("var", "wtn", "n", "prop", "base_w", "count", "nam")
table_2 <- rbind(table_2, ti)
}
table_cl_2 <- table_2 %>% filter(!is.na(var)) %>%
mutate(var_id = paste0(nam, ".", var))
table_cl_compare <- merge(table_cl_1, table_cl_2, by = "var_id", all = T)
names(table_cl_compare) <- c("var.id", "var.hyp", "wtn.hyp", "n.hyp", "prop.hyp", "base.hyp", "base_nw.hyp","nam.hyp", "var.2", "wtn.2", "n.2", "prop.2", "base.2", "base_nw.2", "nam.2")
#Dealing with NA issues
table_cl_compare$nam.hyp[is.na(table_cl_compare$nam.hyp)] <- table_cl_compare$nam.2[is.na(table_cl_compare$nam.hyp)]
table_cl_compare$nam.2[is.na(table_cl_compare$nam.2)] <- table_cl_compare$nam.hyp[is.na(table_cl_compare$nam.2)]
table_cl_compare$to_split <- table_cl_compare$nam.hyp
table_cl_compare <- separate(table_cl_compare, col = to_split, into = c("indctr", "item"), sep ="\\.")
table_cl_compare <- table_cl_compare %>% group_by(indctr) %>% fill(base_nw.hyp, .direction = "down") %>% fill(base_nw.hyp, .direction = "up") %>% ungroup()
table_cl_compare <- table_cl_compare %>% group_by(indctr) %>% fill(base.hyp, .direction = "down") %>% fill(base.hyp, .direction = "up") %>% ungroup()
table_cl_compare <- table_cl_compare %>% group_by(indctr) %>% fill(base_nw.2, .direction = "down") %>% fill(base_nw.2, .direction = "up") %>% ungroup()
table_cl_compare <- table_cl_compare %>% group_by(indctr) %>% fill(base.2) %>% fill(base_nw.hyp, .direction = "down") %>% fill(base.2, .direction = "up") %>% ungroup()
table_cl_compare$prop.hyp[is.na(table_cl_compare$prop.hyp)] <- 0
table_cl_compare$prop.2[is.na(table_cl_compare$prop.2)] <- 0
table_cl_compare$wtn.hyp[is.na(table_cl_compare$wtn.hyp)] <- 0
table_cl_compare$wtn.2[is.na(table_cl_compare$wtn.2)] <- 0
table_cl_compare$chi2 <- chisq.test(table_cl_compare$wtn.2, p = table_cl_compare$prop.hyp)$statistic
table_cl_compare$p <- chisq.test(table_cl_compare$wtn.2, p = table_cl_compare$prop.hyp)$p.value
table_cl_compare$df <- chisq.test(table_cl_compare$wtn.2, p = table_cl_compare$prop.hyp)$df
table_cl_compare$H0 <- ifelse(table_cl_compare$p <= 0.05, "H1 - different", "H0 - same")
return(table_cl_compare)
}Using the function with a single choice question
chi2_GoF(name = "f11_dinking_water_source", stratum_hyp = "a1_informed_consent", stratum_2 = "strata", value_stratum_hyp = "yes", value_stratum_2 = "5km_rural", data = main_dataset, weights = "stratum.weight")## Warning: Expected 2 pieces. Missing pieces filled with `NA` in 8 rows [1, 2, 3,
## 4, 5, 6, 7, 8].## Warning in chisq.test(table_cl_compare$wtn.2, p = table_cl_compare$prop.hyp):
## Chi-squared approximation may be incorrect
## Warning in chisq.test(table_cl_compare$wtn.2, p = table_cl_compare$prop.hyp):
## Chi-squared approximation may be incorrect
## Warning in chisq.test(table_cl_compare$wtn.2, p = table_cl_compare$prop.hyp):
## Chi-squared approximation may be incorrect## # A tibble: 8 × 20
## var.id var.hyp wtn.hyp n.hyp prop.hyp base.hyp base_nw.hyp nam.hyp var.2
## <chr> <chr> <dbl> <int> <dbl> <dbl> <int> <chr> <chr>
## 1 f11_dinking… bottle… 79.5 68 0.0492 1617. 1617 f11_di… bott…
## 2 f11_dinking… drinki… 425. 366 0.263 1617. 1617 f11_di… drin…
## 3 f11_dinking… other_… 8.39 11 0.00519 1617. 1617 f11_di… othe…
## 4 f11_dinking… person… 271. 414 0.168 1617. 1617 f11_di… pers…
## 5 f11_dinking… public… 152. 187 0.0937 1617. 1617 f11_di… publ…
## 6 f11_dinking… tap_dr… 586. 479 0.362 1617. 1617 f11_di… tap_…
## 7 f11_dinking… techni… 2.97 2 0.00184 1617. 1617 f11_di… <NA>
## 8 f11_dinking… trucke… 92.1 90 0.0569 1617. 1617 f11_di… truc…
## # … with 11 more variables: wtn.2 <dbl>, n.2 <int>, prop.2 <dbl>, base.2 <dbl>,
## # base_nw.2 <int>, nam.2 <chr>, indctr <chr>, item <chr>, chi2 <dbl>,
## # p <dbl>, H0 <chr>Using the function with the multiple responce set
names <- names(main_dataset)
f12_names <- names[grepl("f12_drinking_water_treat.", names)]
#removing empty variables from the MR set
nz_f12 <- non_zero_for_test(f12_names, "strata", main_dataset, per_cell = F)## 2 variables were removed because of containing zeros in stratas##
## 20km_rural 20km_urban 5km_rural 5km_urban
## 0 396 389 395 387
## 1 11 15 7 17f12_sig2 <- lapply(nz_f12, chi2_GoF, stratum_hyp = "a1_informed_consent", stratum_2 = "strata", value_stratum_hyp = "yes", value_stratum_2 = "5km_rural", data = main_dataset, weights = "stratum.weight")
f12_sig2_df <- Reduce(rbind, f12_sig2)
f12_sig2_df## # A tibble: 16 × 20
## var.id var.hyp wtn.hyp n.hyp prop.hyp base.hyp base_nw.hyp nam.hyp var.2
## <chr> <int> <dbl> <int> <dbl> <dbl> <int> <chr> <int>
## 1 f12_drinki… 0 1614. 1615 0.998 1617. 1617 f12_dr… 0
## 2 f12_drinki… 1 3.23 2 0.00200 1617. 1617 f12_dr… NA
## 3 f12_drinki… 0 1420. 1437 0.878 1617. 1617 f12_dr… 0
## 4 f12_drinki… 1 197. 180 0.122 1617. 1617 f12_dr… 1
## 5 f12_drinki… 0 1512. 1518 0.935 1617. 1617 f12_dr… 0
## 6 f12_drinki… 1 105. 99 0.0651 1617. 1617 f12_dr… 1
## 7 f12_drinki… 0 1559. 1567 0.964 1617. 1617 f12_dr… 0
## 8 f12_drinki… 1 58.3 50 0.0360 1617. 1617 f12_dr… 1
## 9 f12_drinki… 0 1222. 1226 0.756 1617. 1617 f12_dr… 0
## 10 f12_drinki… 1 395. 391 0.244 1617. 1617 f12_dr… 1
## 11 f12_drinki… 0 1613. 1611 0.998 1617. 1617 f12_dr… 0
## 12 f12_drinki… 1 3.69 6 0.00228 1617. 1617 f12_dr… 1
## 13 f12_drinki… 0 1614. 1614 0.998 1617. 1617 f12_dr… 0
## 14 f12_drinki… 1 2.86 3 0.00177 1617. 1617 f12_dr… NA
## 15 f12_drinki… 0 660. 624 0.408 1617. 1617 f12_dr… 0
## 16 f12_drinki… 1 957. 993 0.592 1617. 1617 f12_dr… 1
## # … with 11 more variables: wtn.2 <dbl>, n.2 <int>, prop.2 <dbl>, base.2 <dbl>,
## # base_nw.2 <int>, nam.2 <chr>, indctr <chr>, item <chr>, chi2 <dbl>,
## # p <dbl>, H0 <chr>7.10.5 Proportion tests (z-test)
Test for equality of proportions for independent samples. To compare column proportions for independent samples, two tailed z-test of proportions could be used.
Main assumptions of use of this test are:
- normally distributed data
- sample size > 30 and n at each point >= 5 (!Thus, for the variables were there are few observations, approximation could be incorrect!) The detailed explanation of this statistic and its use in R could be found here: http://www.sthda.com/english/wiki/two-proportions-z-test-in-r
As in our datasets weighting is used, in the current test, we use weighted proportions and weighted counts for calculating test statistics.
zprop_compare_weighted <- function(data_old, data_new, stratum, value_stratum, name, weights, strict = F){
names_new <- names(data_new)
not_in_new <- names_new[which(!(name %in% names_new))]
i <- 1
table_old <- data.frame()
for (i in 1:length(name)) {
ni <- name[i]
ti <- data_old %>%
group_by(!!sym(ni)) %>% filter(!!sym(stratum) == paste0(value_stratum)) %>%
filter(!is.na(!!sym(ni))) %>%
filter(!!sym(ni) != 'refuse') %>%
dplyr::summarize(wtn = sum(!!sym(weights)), n = n()) %>%
mutate(prop = wtn/sum(wtn), base = sum(wtn), cnt = sum(n))%>%
mutate(varnam = ni)
names(ti) <- c("var", "wtn", "n", "prop", "base_w", "count", "nam")
table_old <- rbind(table_old, ti)
}
table_cl_old <- table_old %>% filter(var != "0") %>% filter(!is.na(var)) %>%
mutate(var_id = paste0(nam, ".", var))
i <- 1
table_new <- data.frame()
for (i in 1:length(name)) {
ni <- name[i]
ti <- data_new %>%
group_by(!!sym(ni)) %>% filter(!!sym(stratum) == paste0(value_stratum)) %>%
filter(!is.na(!!sym(ni))) %>%
filter(!!sym(ni) != 'refuse') %>%
dplyr::summarize(wtn = sum(!!sym(weights)), n = n()) %>%
mutate(prop = wtn/sum(wtn), base = sum(wtn), cnt = sum(n))%>%
mutate(varnam = ni)
names(ti) <- c("var", "wtn", "n", "prop", "base_w", "count", "nam")
table_new <- rbind(table_new, ti)
}
table_cl_new <- table_new %>% filter(var != "0") %>% filter(!is.na(var)) %>%
mutate(var_id = paste0(nam, ".", var))
table_cl_compare <- merge(table_cl_old, table_cl_new, by = "var_id", all = T)
names(table_cl_compare) <- c("var.id", "var.old", "wtn.old", "n.old", "prop.old", "base.old", "base_nw.old","nam.old", "var.new", "wtn.new", "n.new", "prop.new", "base.new", "base_nw.new", "nam.new")
#Dealing with NA issues
table_cl_compare$nam.old[is.na(table_cl_compare$nam.old)] <- table_cl_compare$nam.new[is.na(table_cl_compare$nam.old)]
table_cl_compare$nam.new[is.na(table_cl_compare$nam.new)] <- table_cl_compare$nam.old[is.na(table_cl_compare$nam.new)]
table_cl_compare$to_split <- table_cl_compare$nam.old
table_cl_compare <- separate(table_cl_compare, col = to_split, into = c("indctr", "item"), sep ="\\.")
table_cl_compare <- table_cl_compare %>% group_by(indctr) %>% fill(base_nw.old, .direction = "down") %>% fill(base_nw.old, .direction = "up") %>% ungroup()
table_cl_compare <- table_cl_compare %>% group_by(indctr) %>% fill(base.old, .direction = "down") %>% fill(base.old, .direction = "up") %>% ungroup()
table_cl_compare <- table_cl_compare %>% group_by(indctr) %>% fill(base_nw.new, .direction = "down") %>% fill(base_nw.new, .direction = "up") %>% ungroup()
table_cl_compare <- table_cl_compare %>% group_by(indctr) %>% fill(base.new) %>% fill(base_nw.old, .direction = "down") %>% fill(base.new, .direction = "up") %>% ungroup()
table_cl_compare$prop.old[is.na(table_cl_compare$prop.old)] <- 0
table_cl_compare$prop.new[is.na(table_cl_compare$prop.new)] <- 0
table_cl_compare$wtn.old[is.na(table_cl_compare$wtn.old)] <- 0
table_cl_compare$wtn.new[is.na(table_cl_compare$wtn.new)] <- 0
for (i in 1:nrow(table_cl_compare)){
pt <- prop.test(x = c(table_cl_compare$wtn.old[i], table_cl_compare$wtn.new[i]), n = c(table_cl_compare$base.old[i], table_cl_compare$base.new[i]))
table_cl_compare$prp_tst_p[i] <- pt$p.value
}
table_cl_compare$sig <- ifelse(table_cl_compare$prp_tst_p < 0.05, "Significant diff.", "Not significant")
table_cl_compare$stratum <- paste0(value_stratum)
table_cl_compare_final <- table_cl_compare %>%
filter(case_when(strict == T ~ (prop.old*n.new >= 10 & prop.old*n.old >=10),
strict == F ~ (base_nw.old >= 30 & base_nw.new >=30)))
table_cl_compare_final <- table_cl_compare_final %>% filter(!(nam.old %in% not_in_new))
if(nrow(table_cl_compare_final) < nrow(table_cl_compare)){
ss_vars <- table_cl_compare$var.id[which(!(table_cl_compare$var.id %in% table_cl_compare_final$var.id))]
warning(paste0("Too small sample size. Variables that were removed from the analysis", ss_vars))
}
return(table_cl_compare_final)
}7.10.5.0.1 Applying the function
Z-tests can be used for wave to wave or round to round comparisons.
Assuming, there was staged data collection and at stage1 Luhansk oblast was assessed, while at stage2 Donetsk oblast was assessed (it is an artificial division, for correct comparison between wave), hence we have two different datasets for Donetska and Luhanska regions (UA14 and UA44 respectively) and the question is whether proportions of female-led households, living in each shelter type are the same.
Thus, H0 - proportions are the same, H1 - proportions are different.
ds_ua14 <- main_dataset |> filter(r1_current_oblast == "UA14")
ds_ua44 <- main_dataset |> filter(r1_current_oblast == "UA44")Applying the function for single choices
- data_old, data_new - dataset1 and dataset2;
- stratum - independent variable (should be the same for both datasets), if there is no need for independent variable, variable that have same value across the entire dataset can be used (e.g. “informed consent”);
- value_stratum - independent variable value, that should be used for crosstabulations (e.g. we would like to compare the proportions for certain settlement or household type);
- name - dependent variable name;
- weights - weighting variable;
- strict - (TRUE/FALSE) specifies if the less strict restrictment to sample size (n>30) or more strict one (n at each point >= 5) should be applied. FALSE be default
library(dplyr)
# without independent variable
marital_prop_test <- zprop_compare_weighted(data_old = ds_ua14, data_new = ds_ua44, stratum = "a1_informed_consent", value_stratum = "yes", name = "b9_hohh_marital_status", weights = "stratum.weight")
marital_prop_test## # A tibble: 6 × 20
## var.id var.old wtn.old n.old prop.old base.old base_nw.old nam.old var.new
## <chr> <chr> <dbl> <int> <dbl> <dbl> <int> <chr> <chr>
## 1 b9_hohh_m… divorc… 141. 127 0.116 1220. 1168 b9_hoh… divorc…
## 2 b9_hohh_m… married 538. 515 0.441 1220. 1168 b9_hoh… married
## 3 b9_hohh_m… separa… 33.3 32 0.0273 1220. 1168 b9_hoh… separa…
## 4 b9_hohh_m… single 113. 99 0.0924 1220. 1168 b9_hoh… single
## 5 b9_hohh_m… unmarr… 61.6 65 0.0505 1220. 1168 b9_hoh… unmarr…
## 6 b9_hohh_m… widowed 333. 330 0.273 1220. 1168 b9_hoh… widowed
## # … with 11 more variables: wtn.new <dbl>, n.new <int>, prop.new <dbl>,
## # base.new <dbl>, base_nw.new <int>, nam.new <chr>, indctr <chr>, item <chr>,
## # prp_tst_p <dbl>, sig <chr>, stratum <chr># with an independent variable
house_type_married <- zprop_compare_weighted(data_old = ds_ua14, data_new = ds_ua44, stratum = "b9_hohh_marital_status", value_stratum = "married", name = "e2_home_type", weights = "stratum.weight")Output numeric columns: - wtn.old - weighted counts for the first (old) data - n.old - unweighted counts for the first (old) data - prop.old - weighted proportion for the first (old) data - base.old - weighted base (total count) for the first (old) data - base_nw.old - unweighted base (total count) for the first (old) data - wtn.new - weighted counts for the second (new) data - n.new - unweighted counts for the second (new) data - prop.new - weighted proportion for for the second (new) data - base.new - weighted base (total count) for the second (new) data - base_nw.new - unweighted base (total count) for the second (new) data - prp_tst_p - p-value for proportions test - sig - indication if p<0.05
Applying the function for multiple choice questions
names <- names(ds_ua14)
ng5 <- names[grepl("b10_hohh_vulnerability", names)]
vulnerability_prop_test <- zprop_compare_weighted(data_old = ds_ua14, data_new = ds_ua44, stratum = "a1_informed_consent", value_stratum = "yes", name = ng5, weights = "stratum.weight")
vulnerability_prop_test## # A tibble: 61 × 20
## var.id var.old wtn.old n.old prop.old base.old base_nw.old nam.old var.new
## <chr> <chr> <dbl> <int> <dbl> <dbl> <int> <chr> <chr>
## 1 b10_hohh… chroni… 75.3 68 0.0618 1220. 1168 b10_ho… chroni…
## 2 b10_hohh… chroni… 7.38 7 0.00605 1220. 1168 b10_ho… chroni…
## 3 b10_hohh… chroni… 16.4 24 0.0134 1220. 1168 b10_ho… chroni…
## 4 b10_hohh… chroni… 0.275 1 0.000225 1220. 1168 b10_ho… <NA>
## 5 b10_hohh… chroni… 10.4 11 0.00855 1220. 1168 b10_ho… chroni…
## 6 b10_hohh… 1 505. 496 0.414 1220. 1168 b10_ho… 1
## 7 b10_hohh… disabi… 24.8 24 0.0204 1220. 1168 b10_ho… disabi…
## 8 b10_hohh… disabi… 12.4 10 0.0102 1220. 1168 b10_ho… disabi…
## 9 b10_hohh… disabi… 4.99 4 0.00410 1220. 1168 b10_ho… <NA>
## 10 b10_hohh… disabi… 2.03 3 0.00167 1220. 1168 b10_ho… <NA>
## # … with 51 more rows, and 11 more variables: wtn.new <dbl>, n.new <int>,
## # prop.new <dbl>, base.new <dbl>, base_nw.new <int>, nam.new <chr>,
## # indctr <chr>, item <chr>, prp_tst_p <dbl>, sig <chr>, stratum <chr>